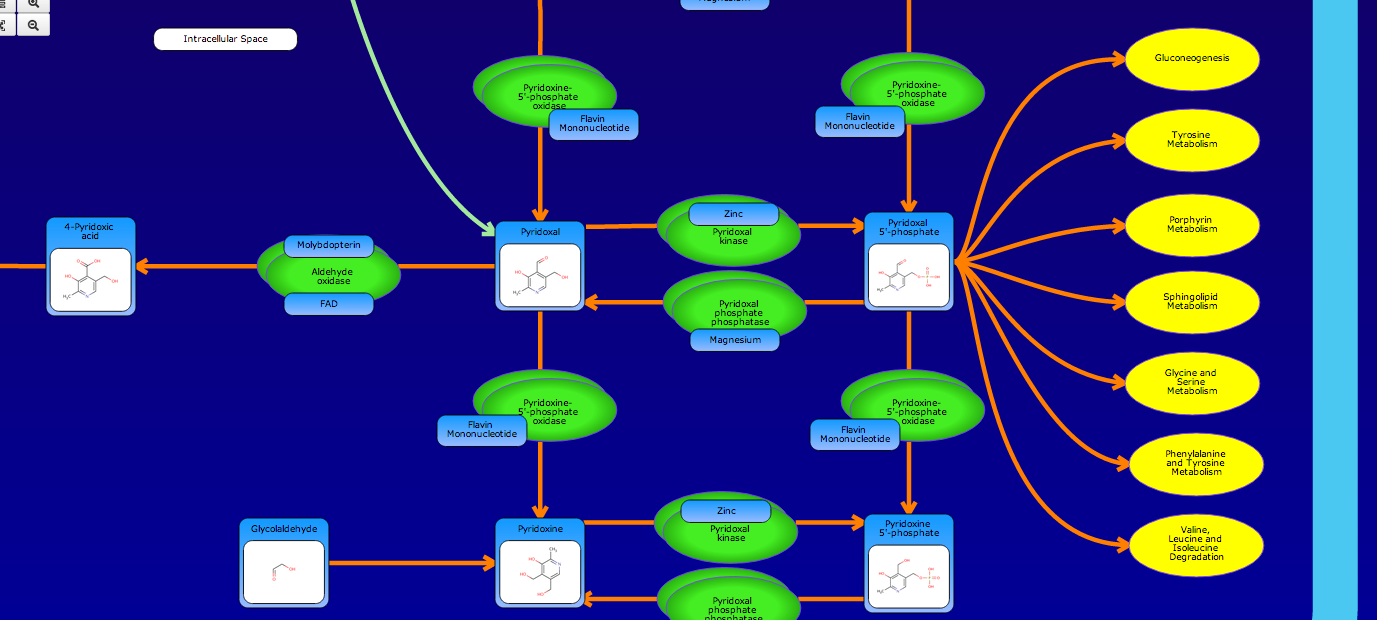
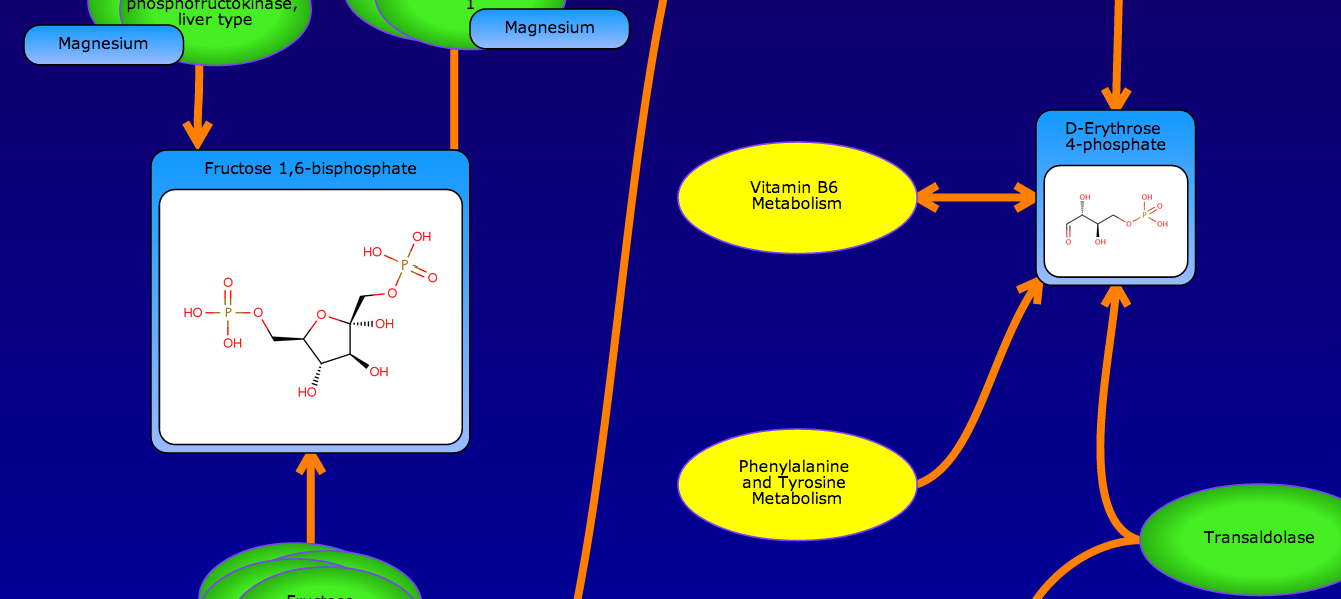
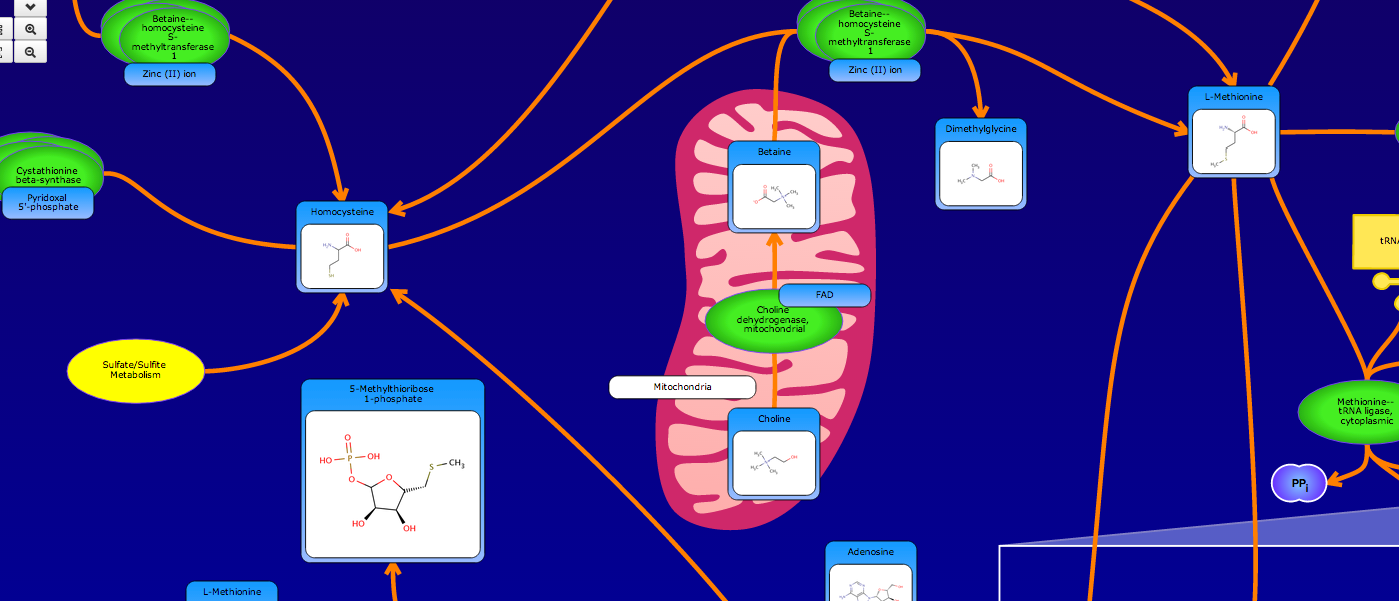
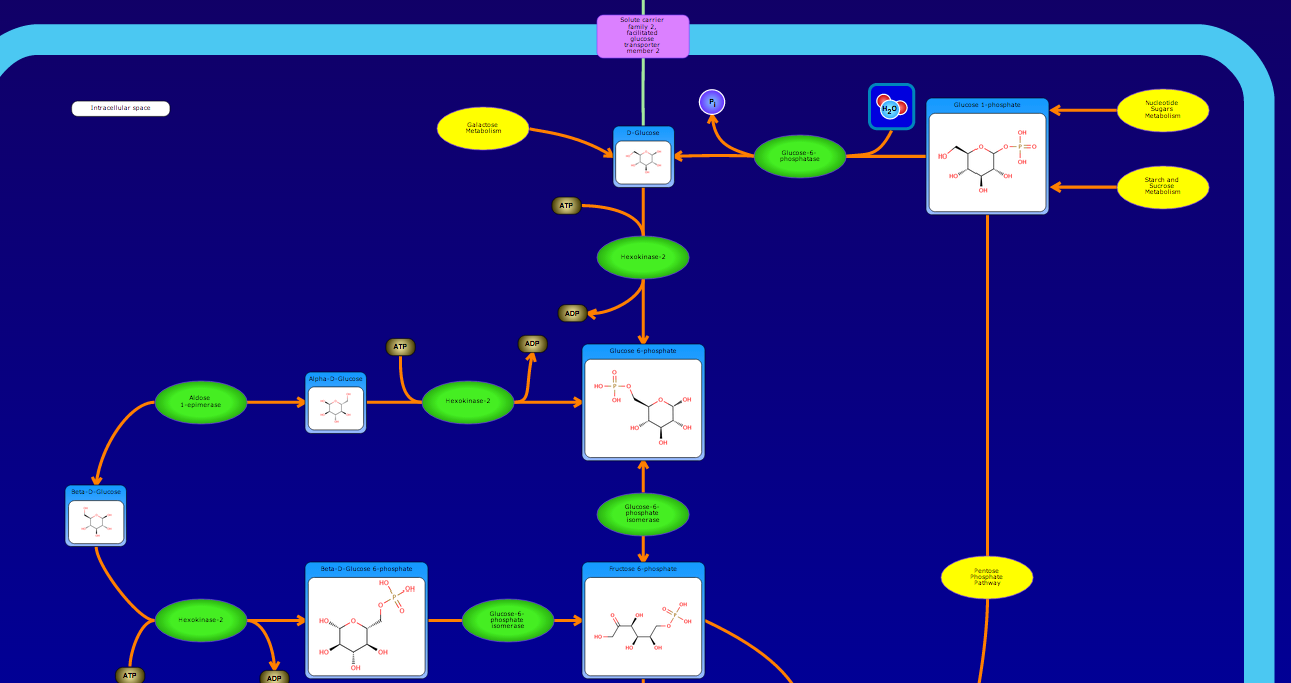
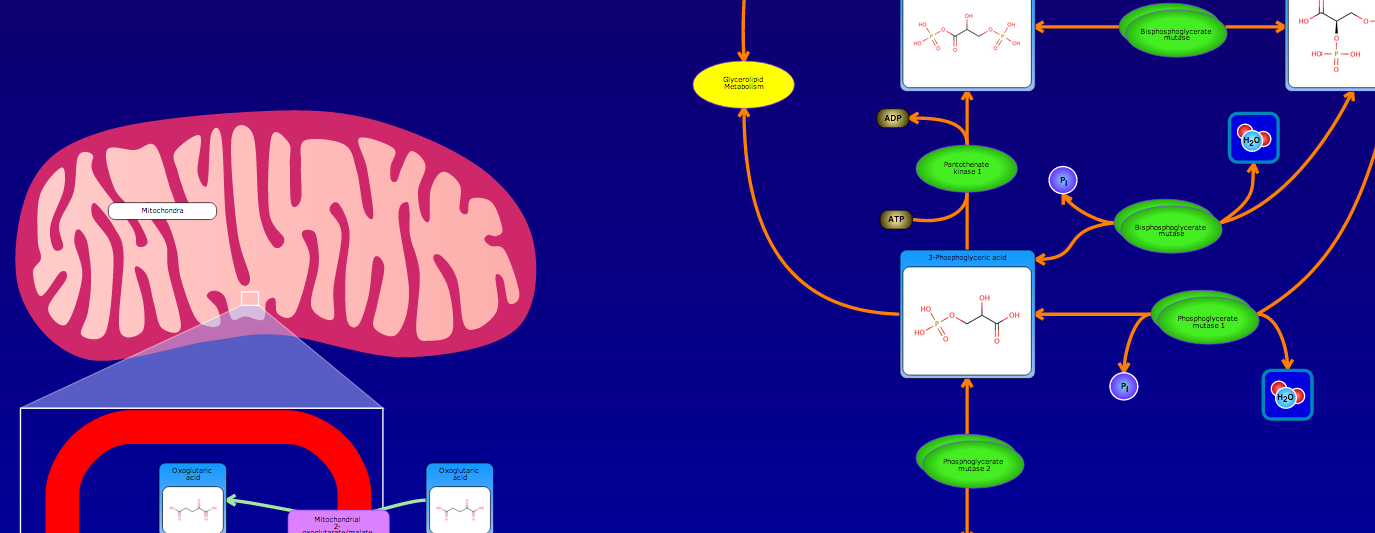
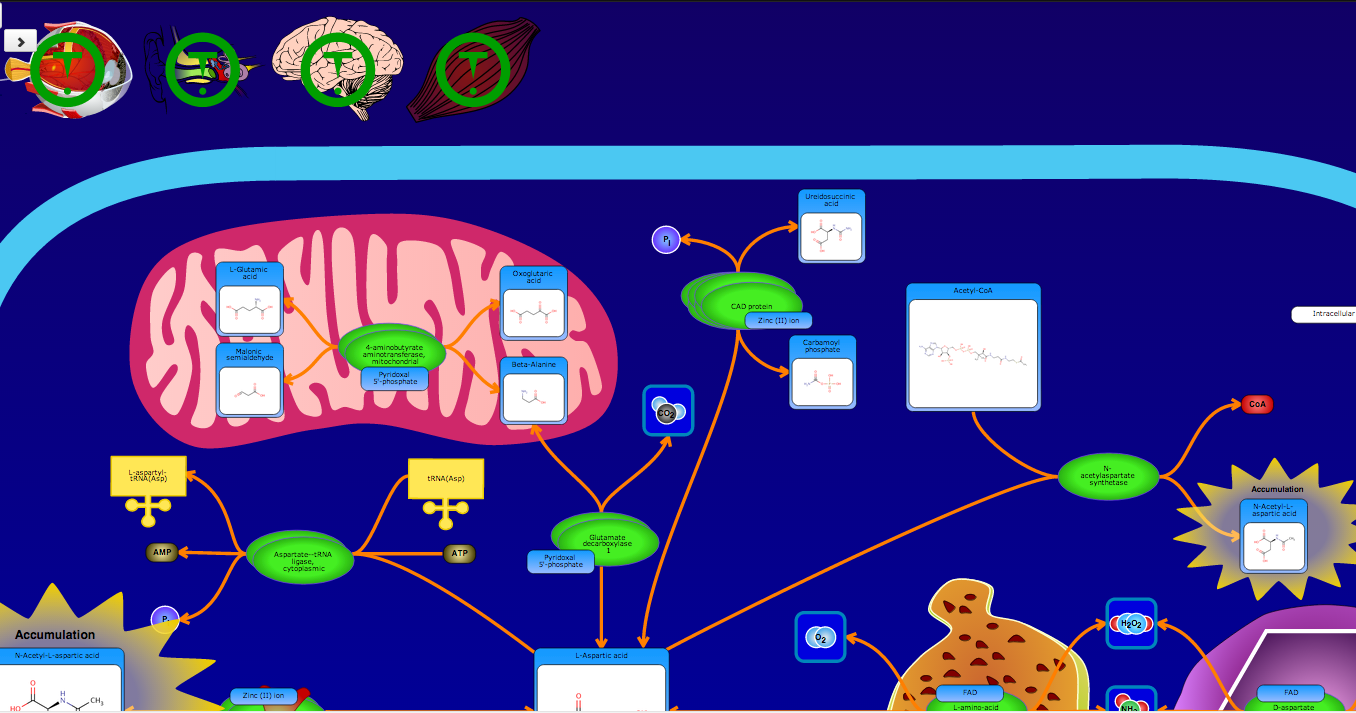
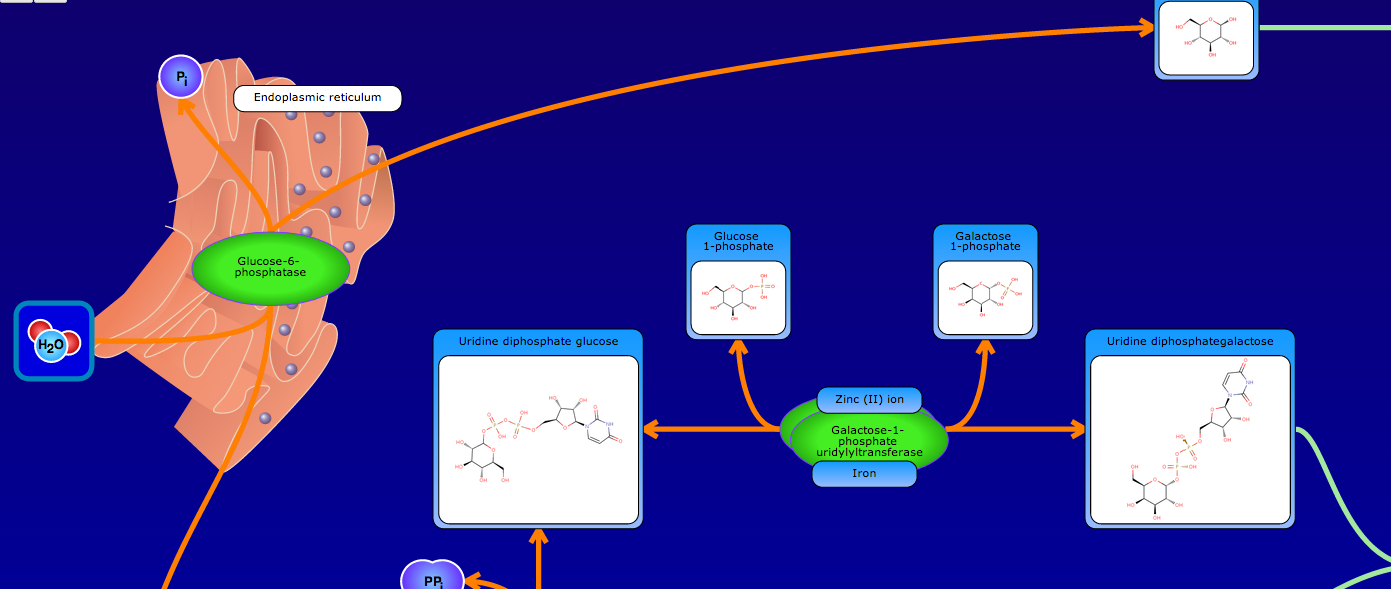
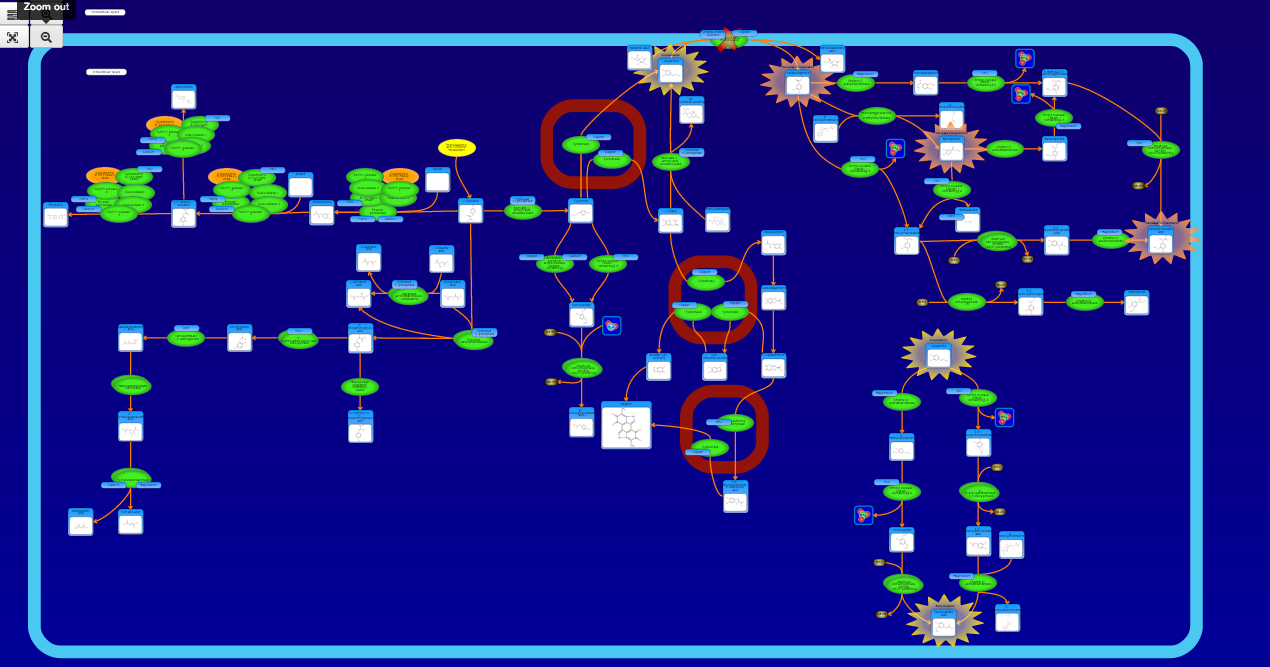
PathBank is an interactive, visual database containing more than 600 000 machine-readable pathways found in model organisms such as humans, mice, E. coli, yeast, and Arabidopsis thaliana. The majority of these pathways are not found in any other pathway database. PathBank is designed specifically to support pathway elucidation and pathway discovery in metabolomics, transcriptomics, proteomics, and systems biology. It is able to do so, in part, by providing detailed, fully searchable, hyperlinked diagrams of metabolic, signalling, disease, drug, and physiological pathways. All PathBank pathways include information on the relevant organelles, subcellular compartments, protein complex cofactors, protein complex locations, metabolite locations, chemical structures, and protein complex quaternary structures. Each small molecule is hyperlinked to detailed descriptions contained in the HMDB or DrugBank and each protein complex or enzyme complex is hyperlinked to UniProt. All PathBank pathways are accompanied with detailed descriptions and references, providing an overview of the pathway, condition, or processes depicted in each diagram. The database is easily browsed and supports full text, sequence, and chemical structure searching. Users may query PathBank with lists of metabolite names, drug names, genes/protein complex names, SwissProt IDs, GenBank IDs, Affymetrix IDs, or Agilent microarray IDs. These queries will produce lists of matching pathways and highlight the matching molecules on each of the pathway diagrams. Gene, metabolite, and protein complex concentration data can also be visualized through PathBank's mapping interface. Each of PathBank's images, descriptions, and tables are downloadable in several formats.
PathBank is supported by David Wishart, Departments of Computing Science & Biological Sciences, University of Alberta. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (PathBank) and the original publication (see below). We ask that users who download significant portions of the database cite the PathBank paper in any resulting publications.
- Wishart DS, Li C, Marcu A, et al. PathBank: A Comprehensive Pathway Database for Model Organisms. Nucleic Acids Res. 2020 Jan 8;48(D1):D470-D478.
- Wishart DS, Kruger R, Sivakumaran A, et al. PathBank 2.0 - The Pathway Database for Model Organism Metabolomics. Nucleic Acids Res. 2024 Jan 5;52(D1):D654-D662.